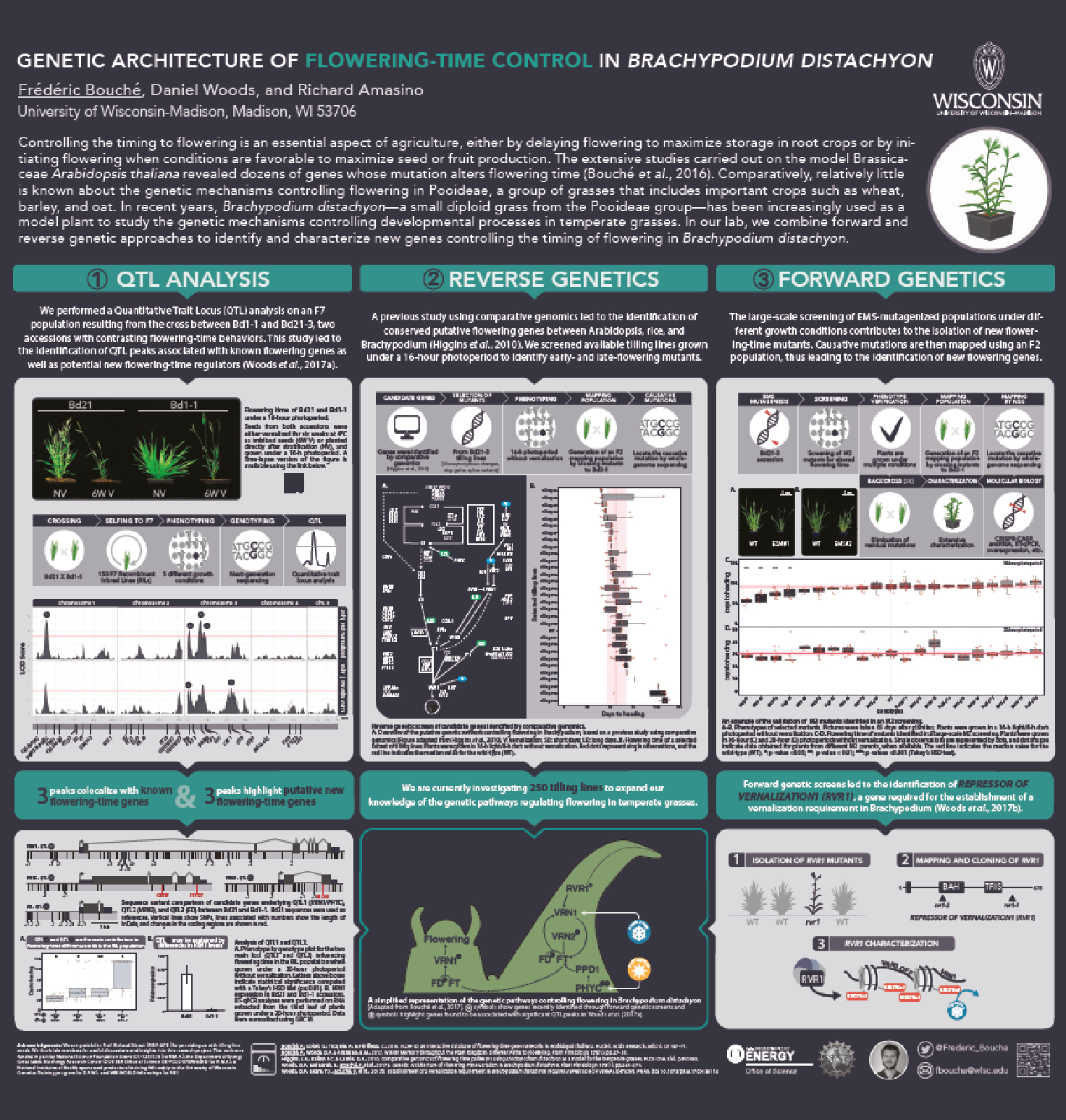
The genetic basis of flowering-time control in Brachypodium
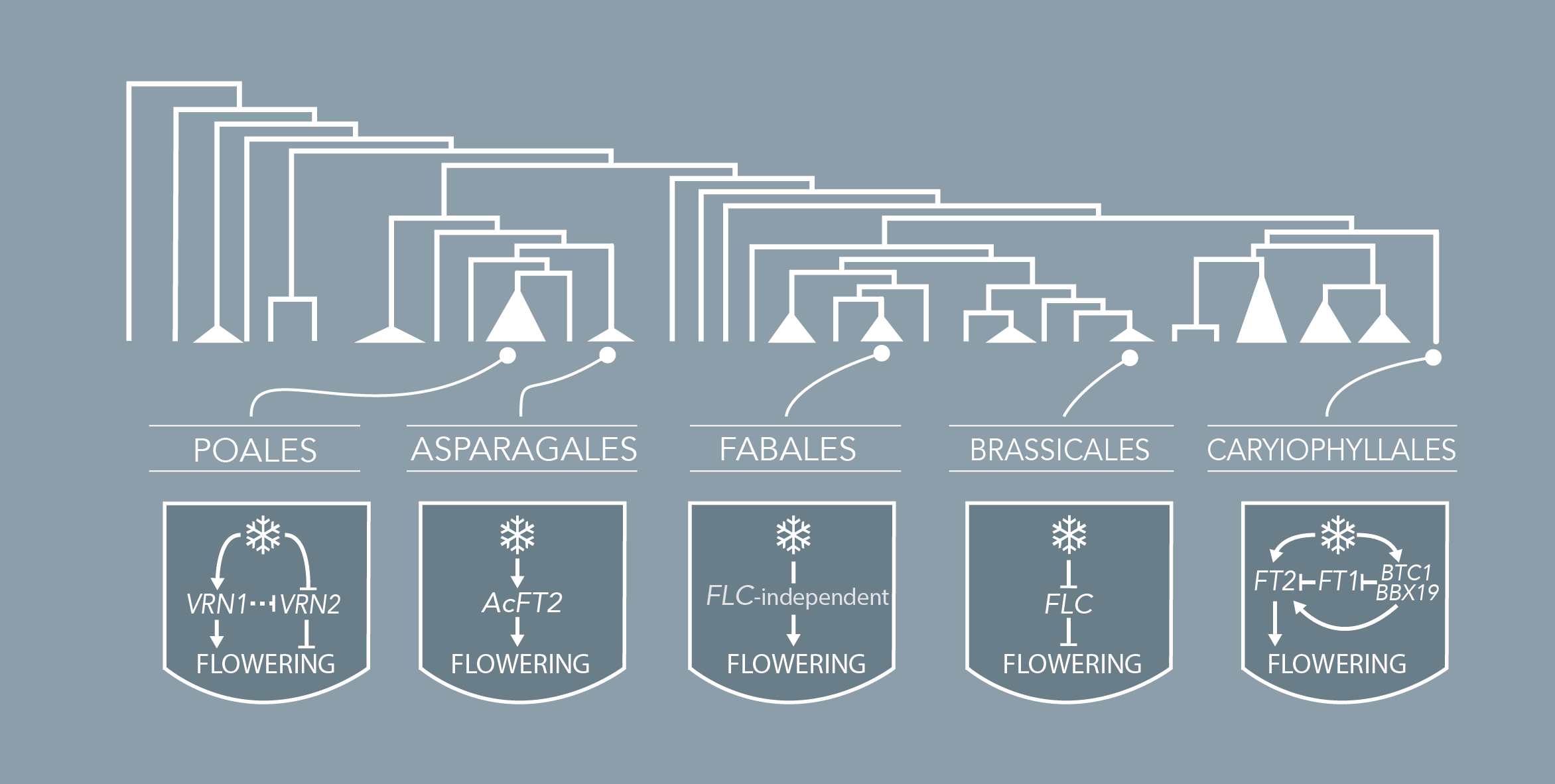
Brachypodium distachyon is a grass species from the Triticeae tribe, which includes wheat, barley, oat, and rye. A better understanding of the molecular mechanisms controlling the initiation of flowering in these species is necessary to improve yields. However, the economically important grasses tend to be physically large, have relatively long life cycles and possess complex genomes, thus making genetic studies very complex. In this context, Brachypodium emerges as a model organism to study the molecular mechanisms governing flowering in those species. The knowledge accumulated on the control of flowering time in Brachypodium will help researchers to better select inbred cereal crops.
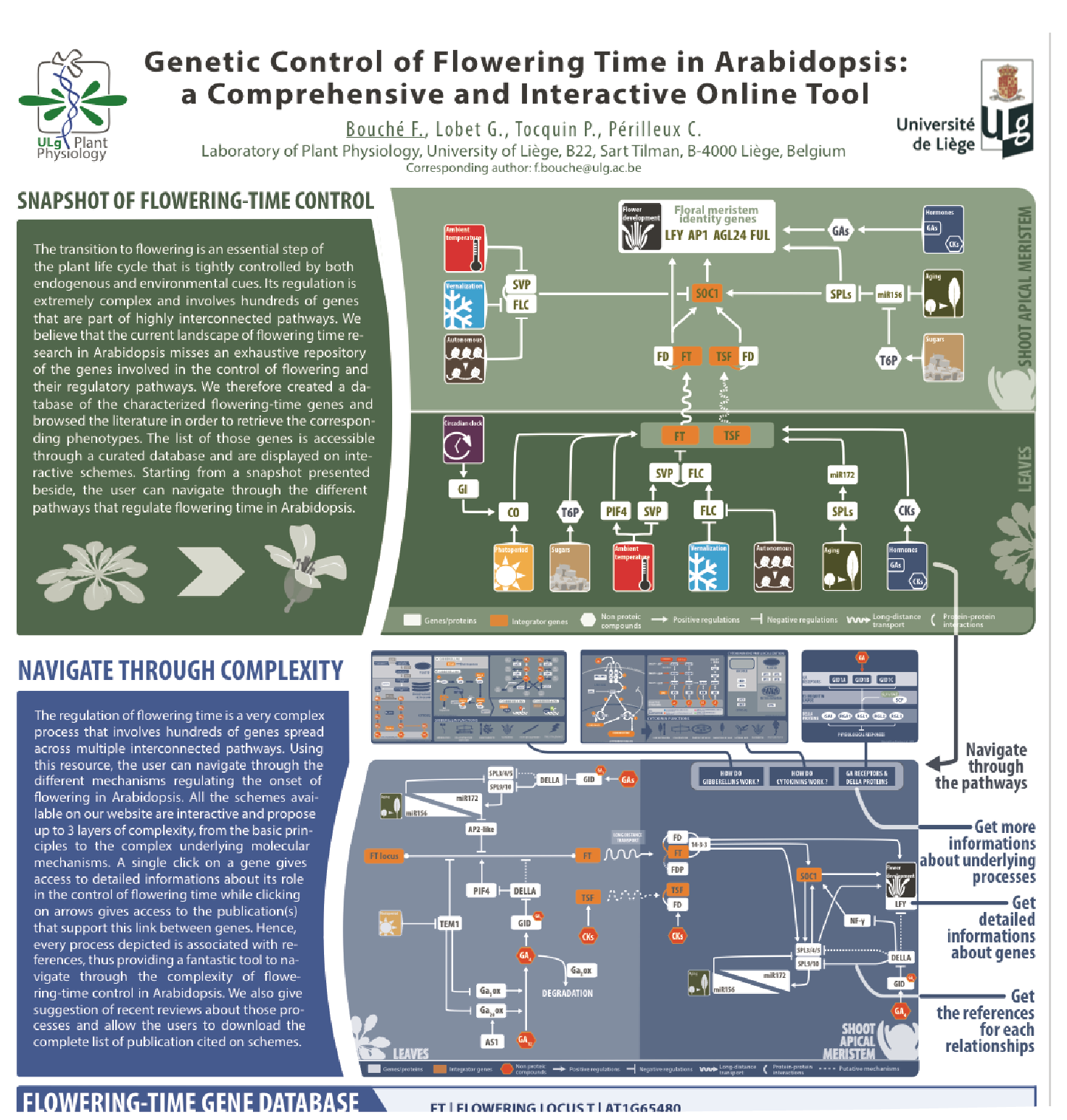
Database of Arabidopsis flowering-time genes
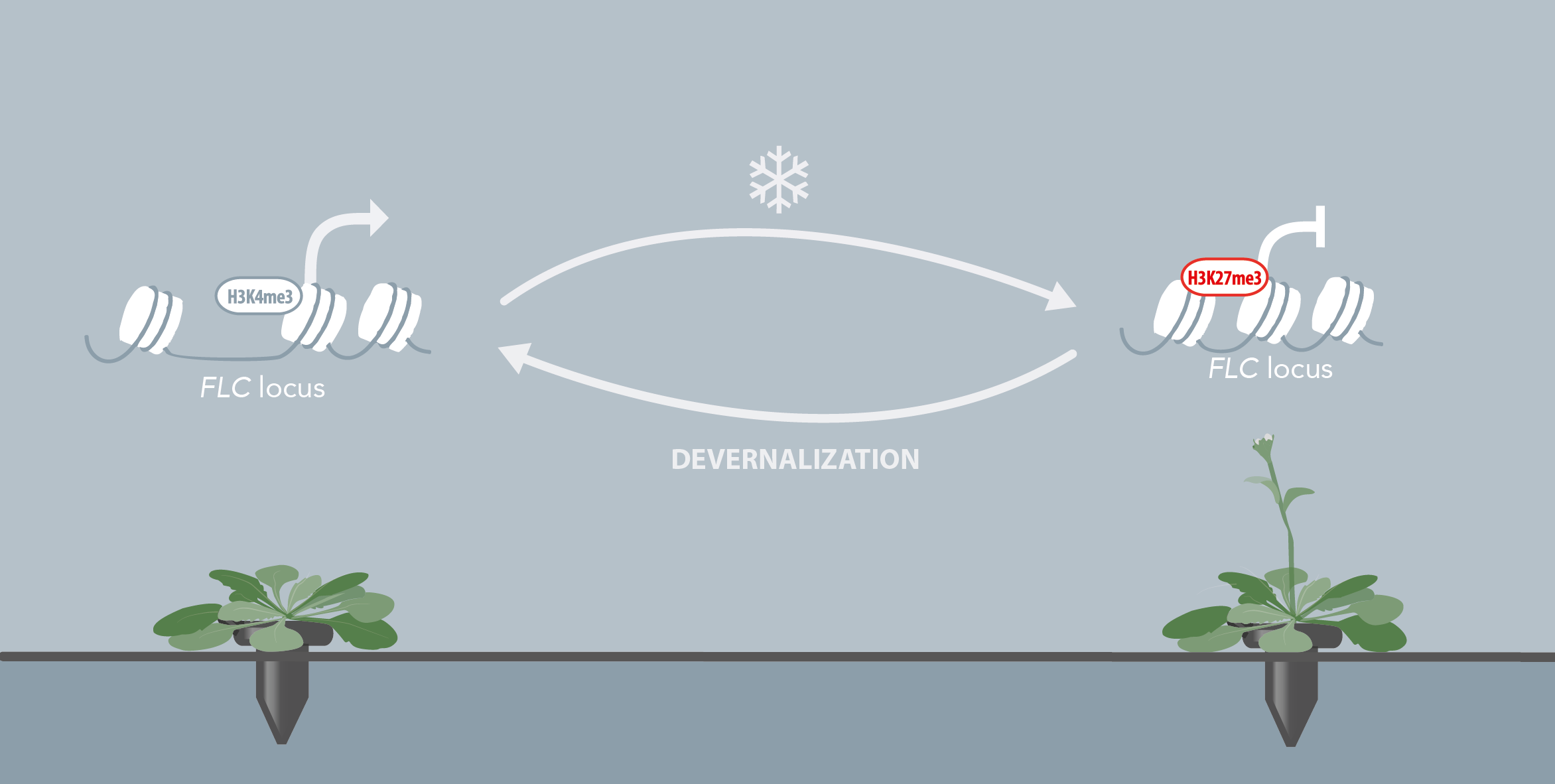
The transition to flowering is an essential step of the plant life cycle that is tightly controlled by both endogenous and environmental cues. Its regulation is extremely complex and involves hundreds of genes that are part of highly interconnected pathways. Our knowledge of the molecular mechanisms governing the floral induction of Arabidopsis thaliana increases quickly and a significant number of reviews are published every year on this topic. We developed a web-based interactive resource organized around a curated database of the flowering time genes, available at www.flor-id.org. This project is a collaboration with Guillaume Lobet, and was published in Nucleic Acids Research.
Read more about it: Flowering highlights - F1000 Prime - ULg website - Video description

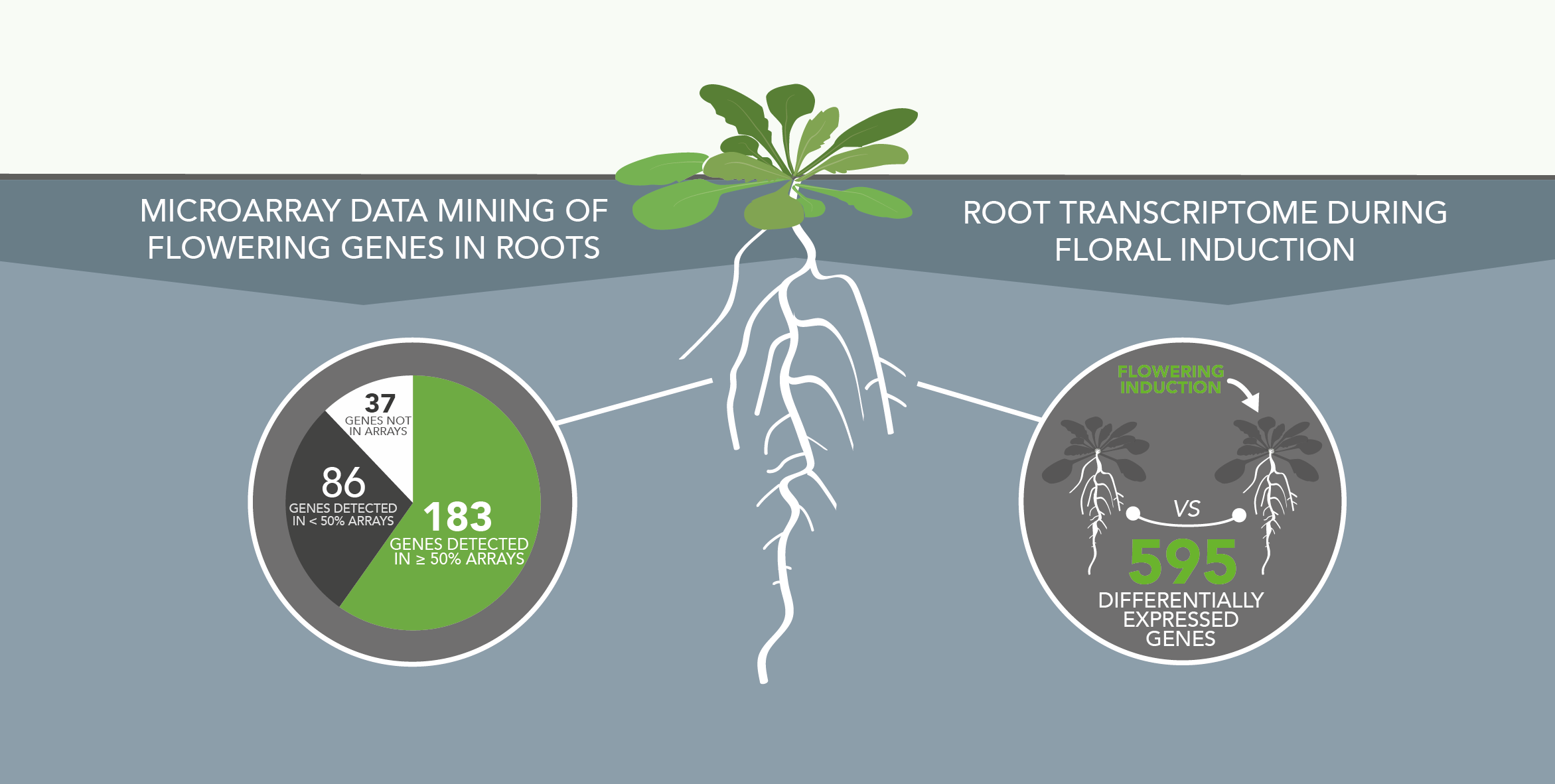
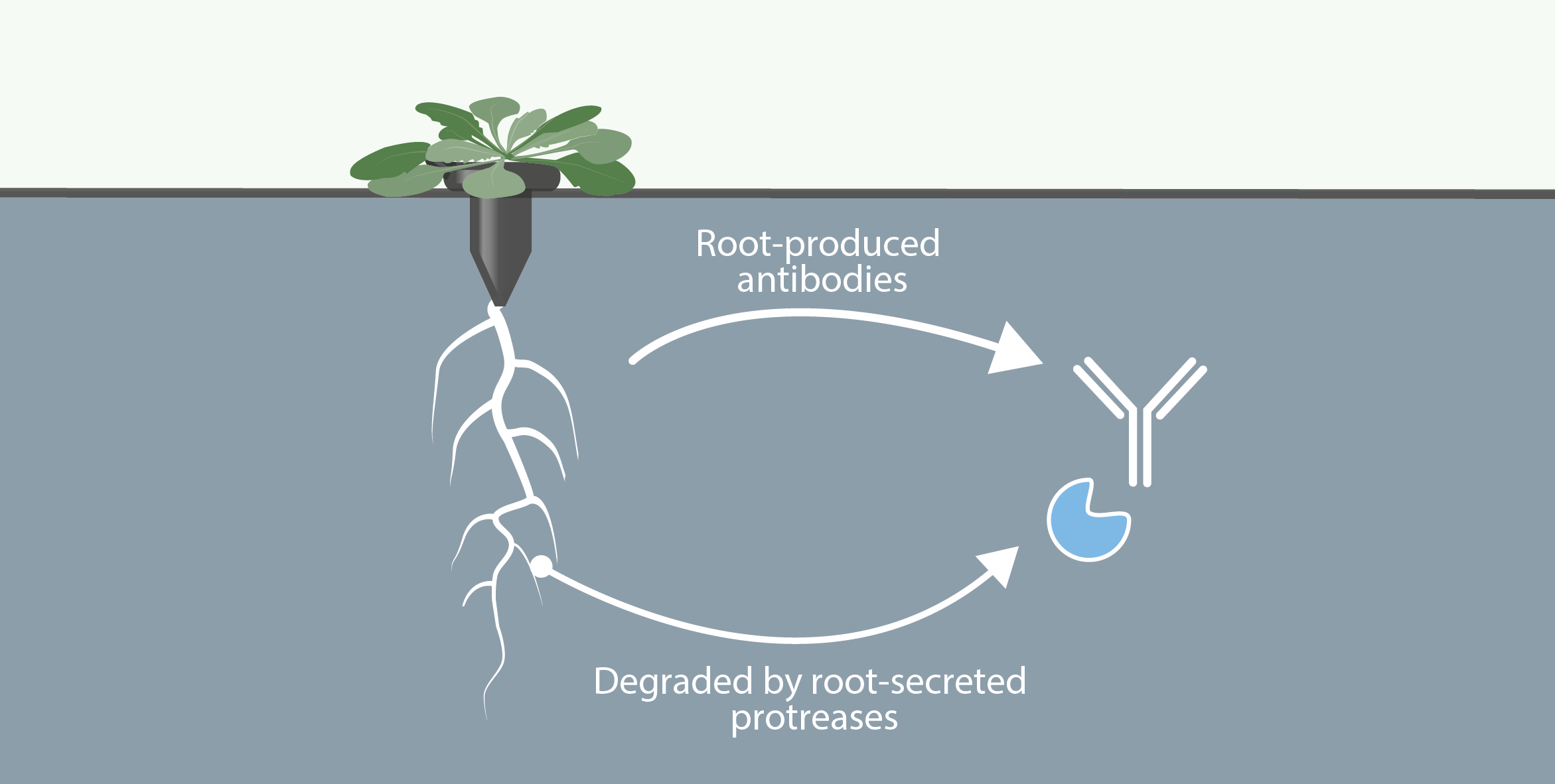
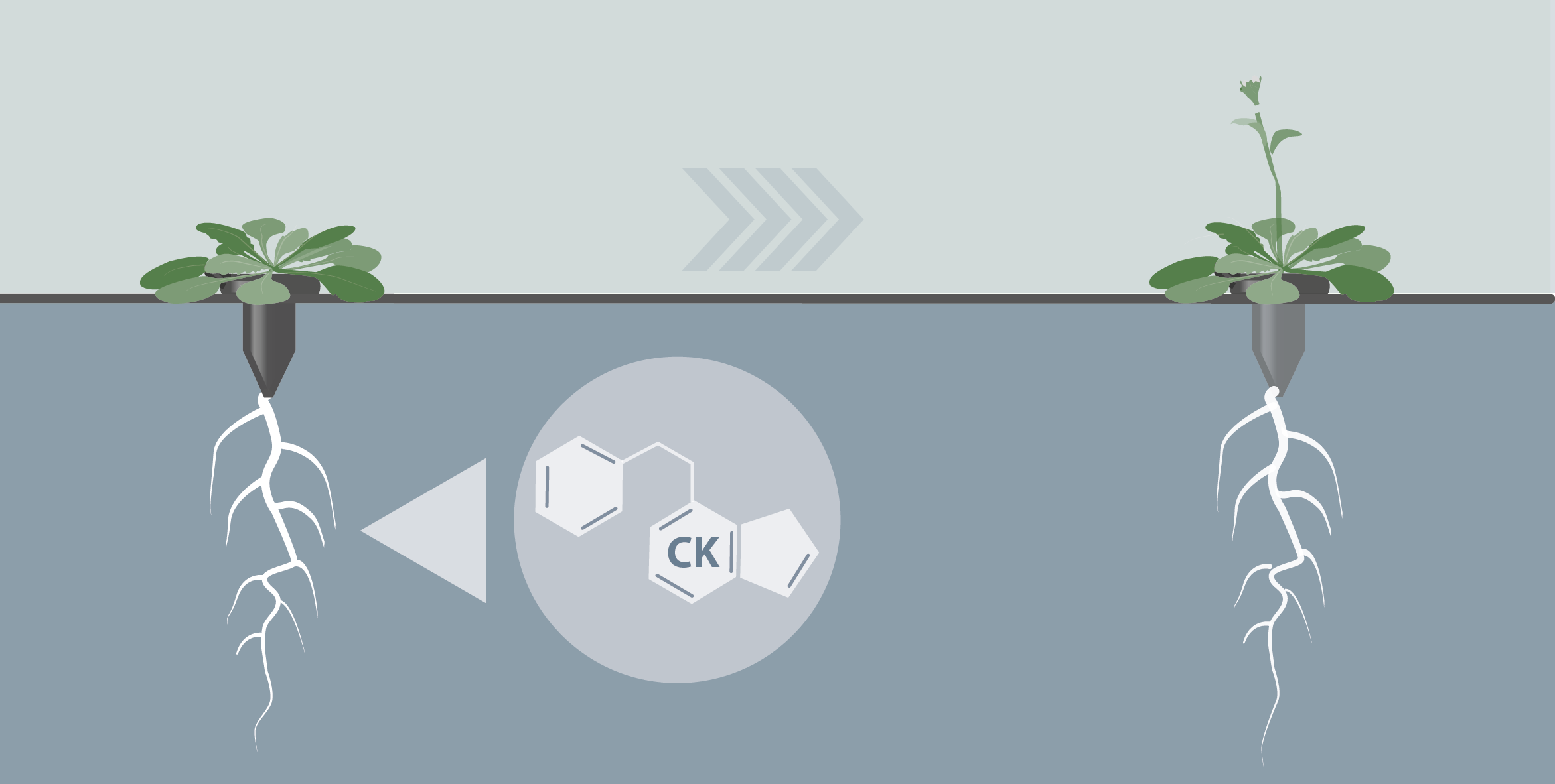
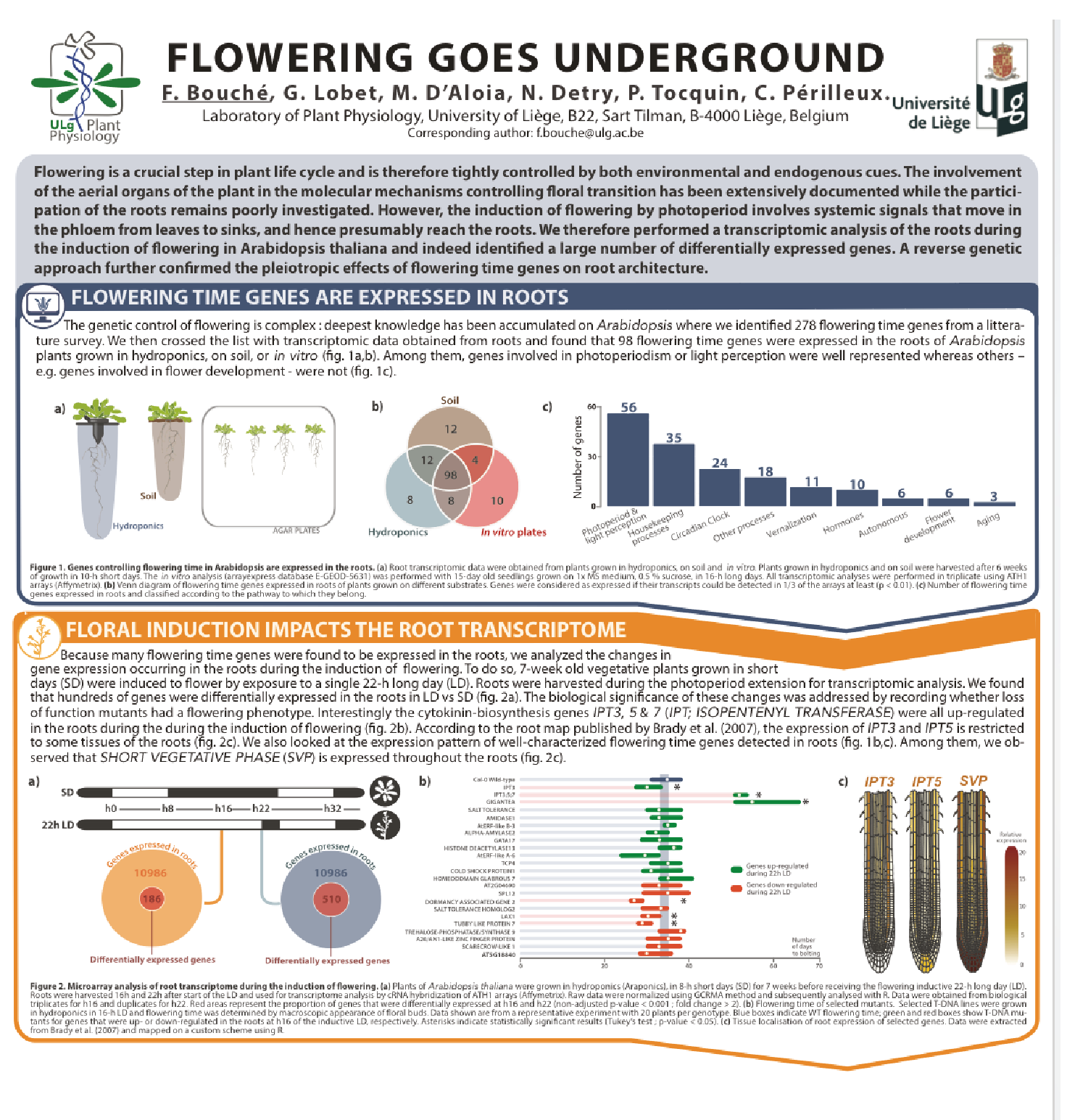
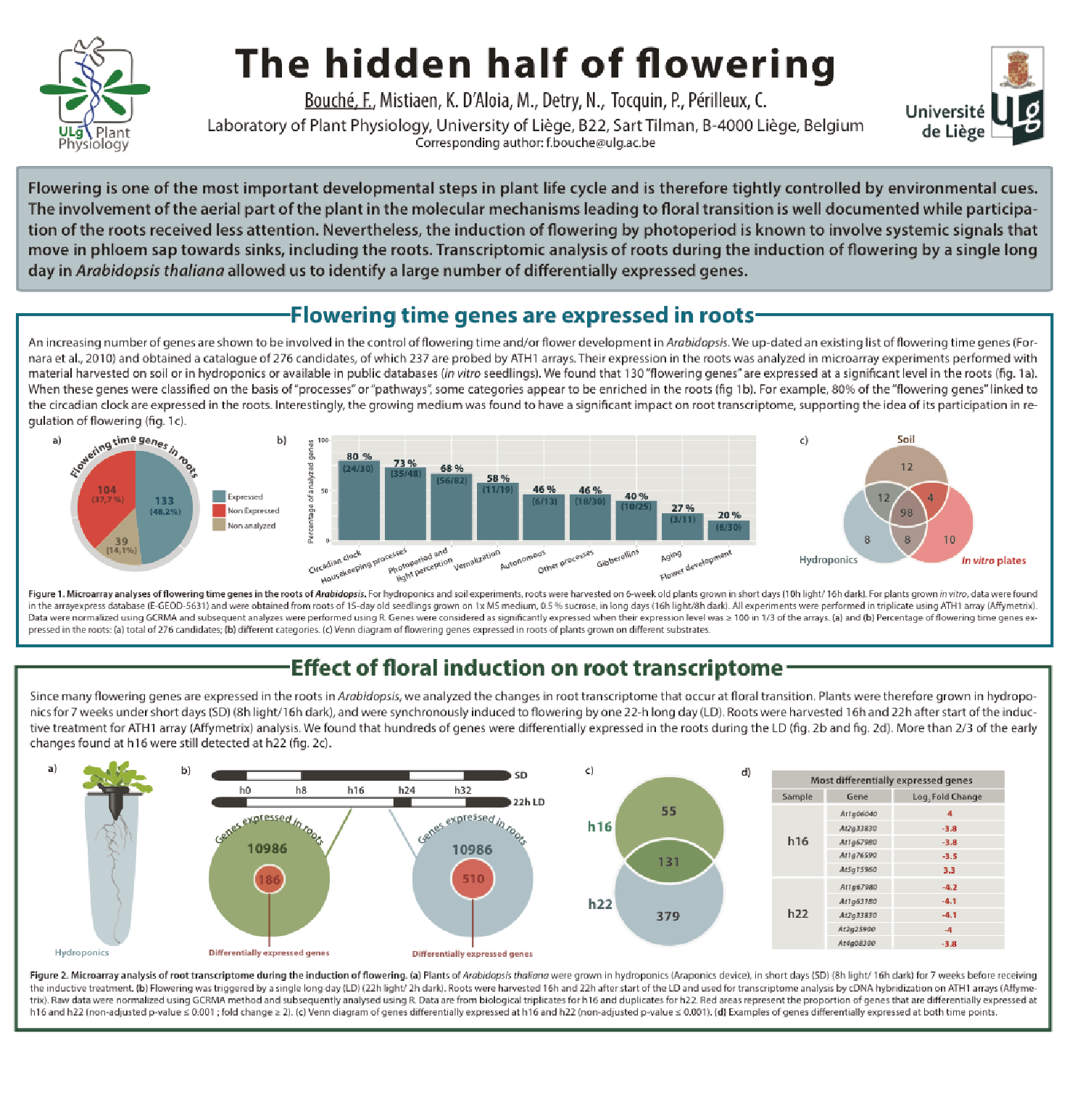
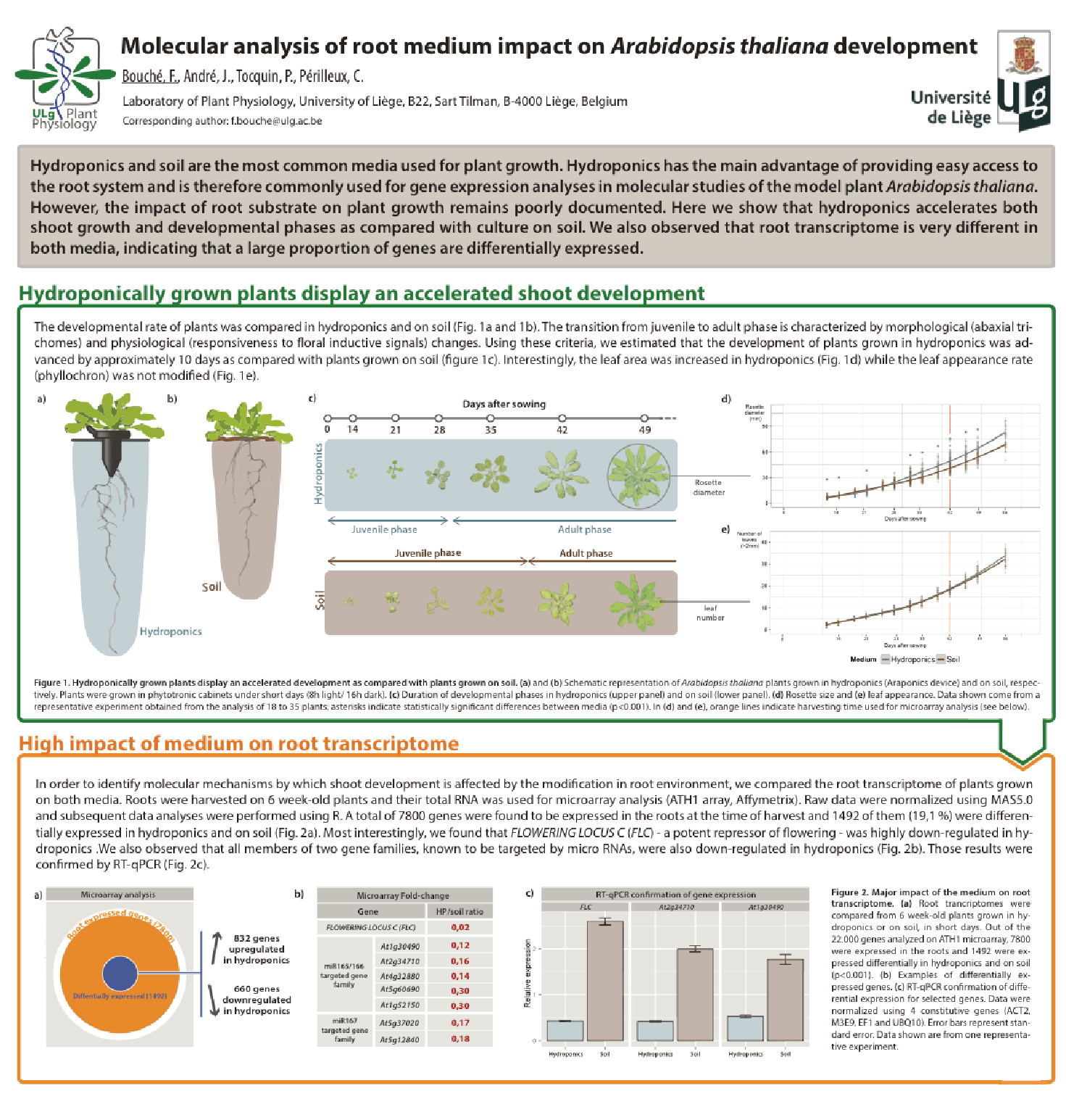
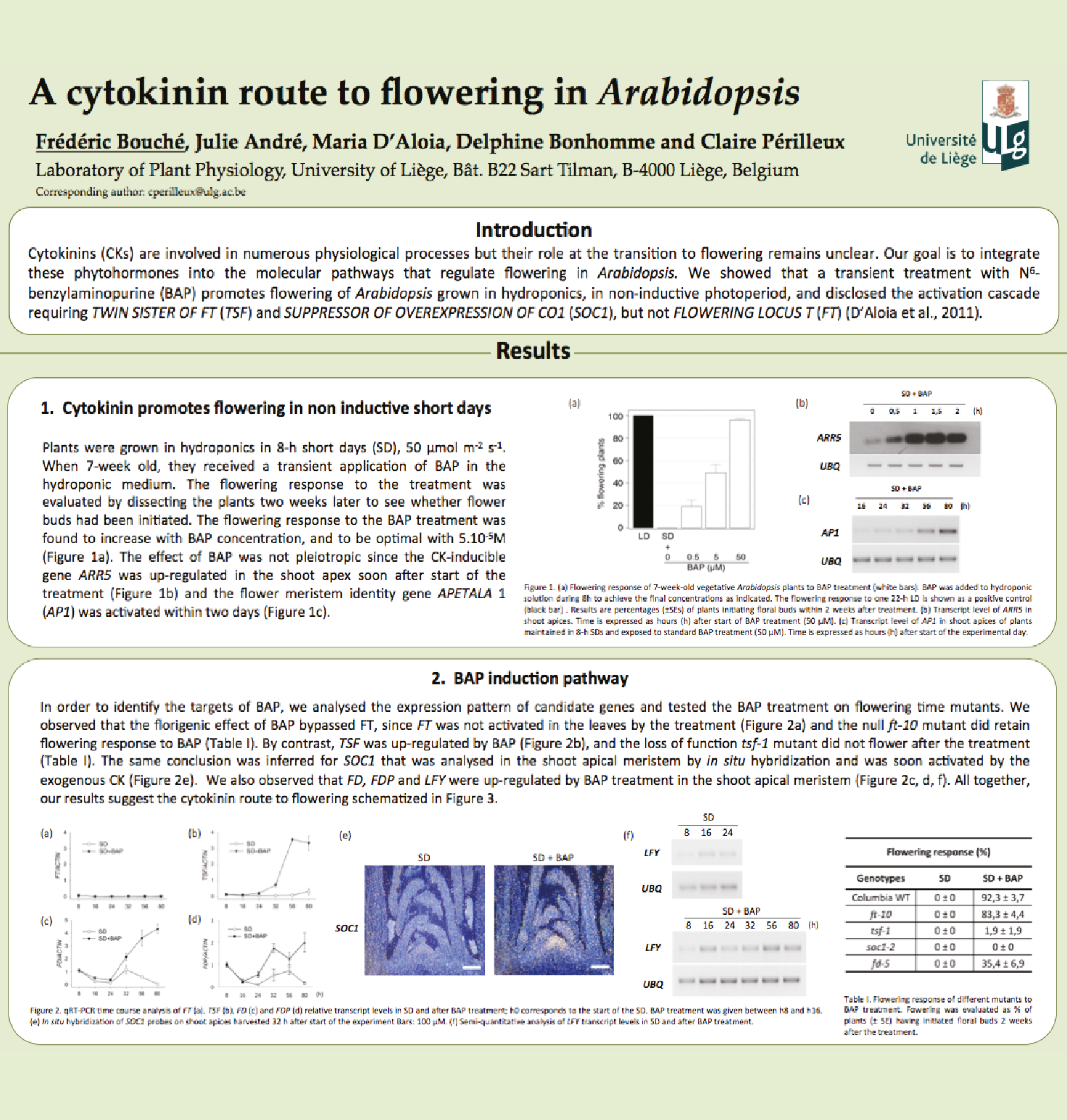
Involvment of roots in floral induction in Arabidopsis
The induction of flowering is a complex process involving hundreds of genes that are part of different interconnected pathways. Together, they converge to regulate a small subset of genes that are responsible for the switch from the vegetative to the reproductive phase of the plant life cycle. Photoperiodic induction of flowering is mediated by a mobile signal produced in phloem’s companion cells that subsequently migrates to the shoot apical meristem and likely reaches roots. My work intends to use an integrative approach to unravel the relationships that exist between roots and shoot during the induction of flowering but also to identify the role of flowering time genes expressed in roots.
PEER-REVIEWED ARTICLES
SELECTED PRESENTATIONS AND POSTERS
MORE ABOUT ME
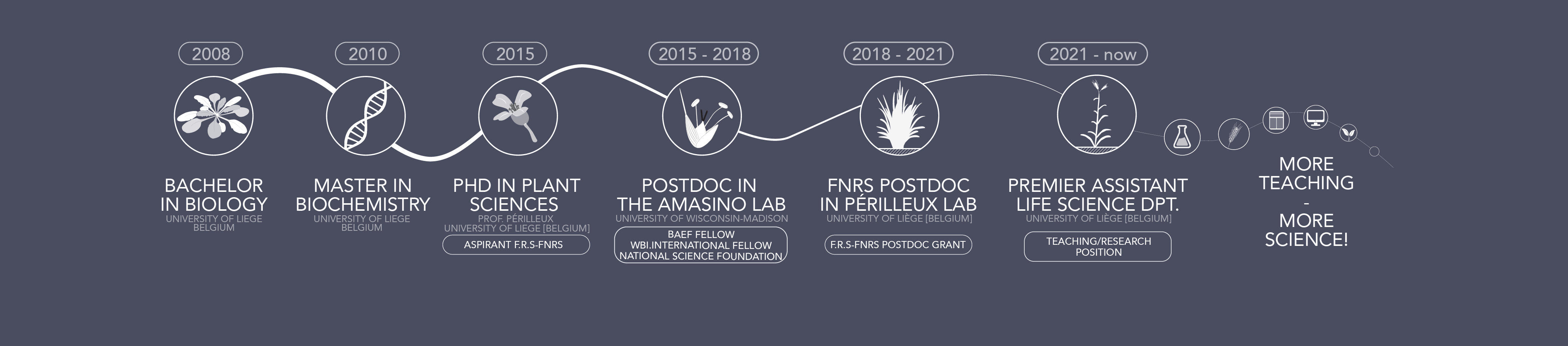
Professionnal background
When I am not doing Science

Funding and Support